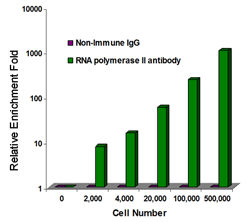
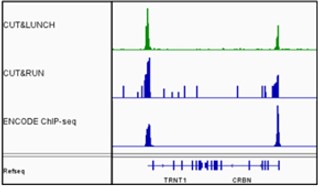
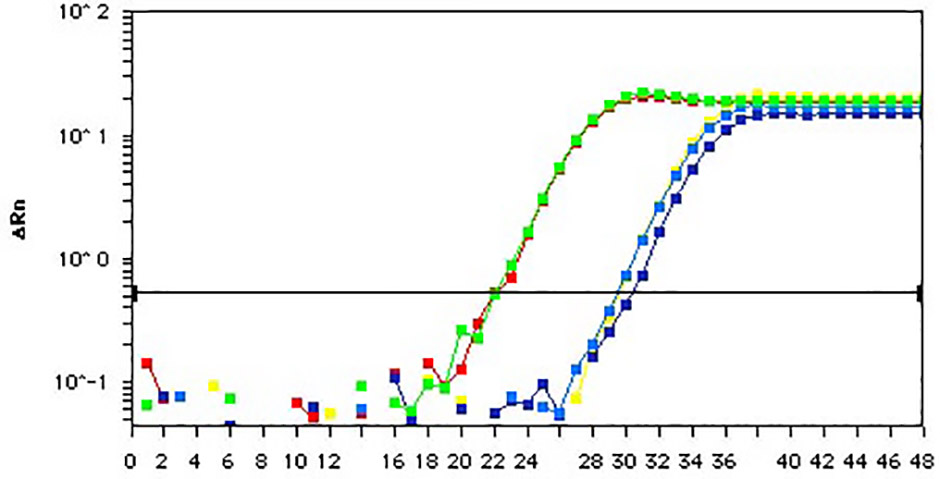
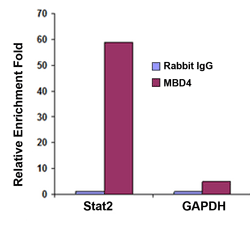
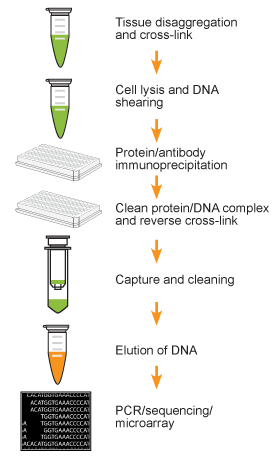
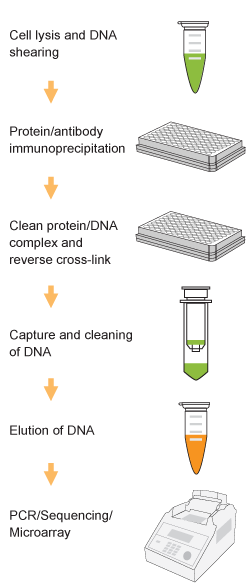
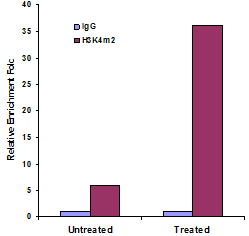
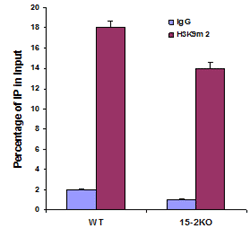
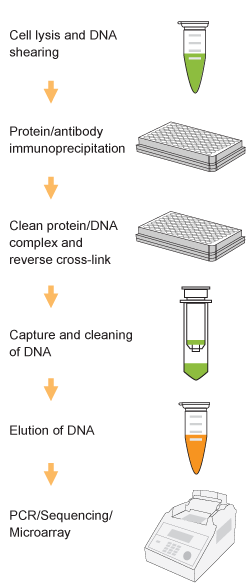
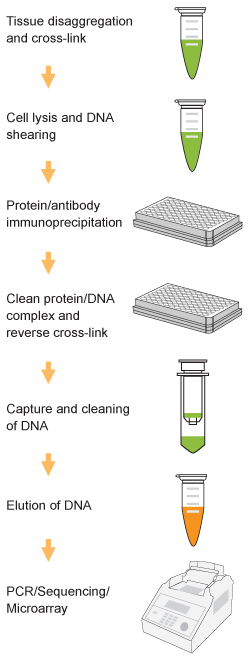
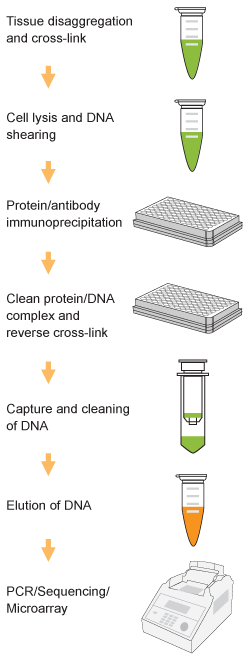
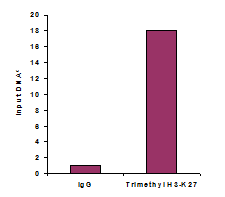Chromatin studies, including chromatin immunoprecipitation (ChIP) and chromatin accessibility assays, are essential for understanding how DNA is packaged and regulated within the nucleus. These techniques help identify protein-DNA interactions, protein post-translational modifications, and accessible regulatory regions that influence gene expression, cell differentiation, and disease states.
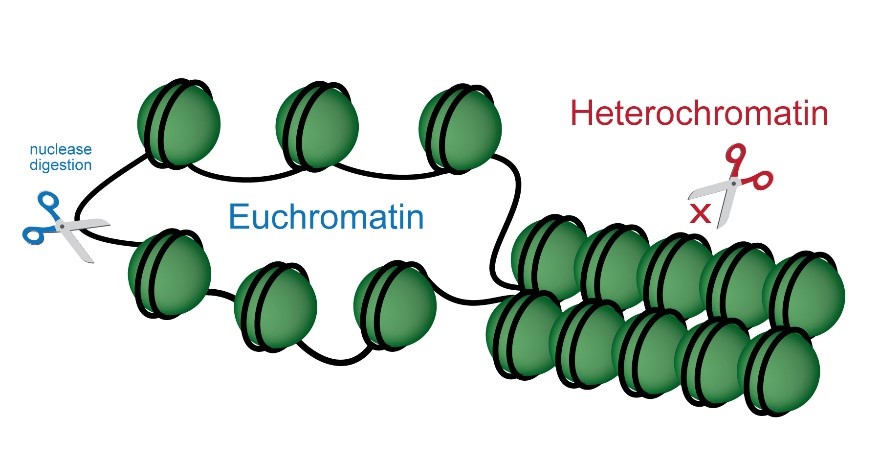
Researchers study chromatin structure and dynamics to uncover mechanisms of gene regulation, epigenetic inheritance, and potential therapeutic targets for diseases.
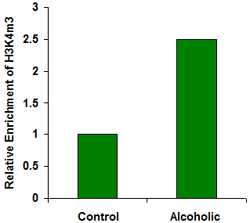
Assess open and closed chromatin configurations gene specifically or genome wide
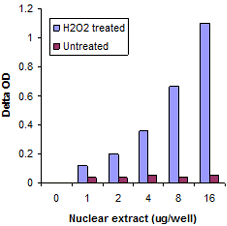
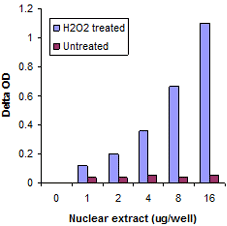
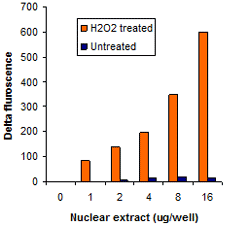
Measure activity and inhibition of HATs, HDACs, HMTs, and HDMTs




 Cart (0)
Cart (0)