Methylation-specific PCR (MSP or MS-PCR) and bisulfite sequencing are techniques used to study the methylation status of cytosine bases in DNA. DNA methylation is a chemical modification that involves the addition of a methyl group to the cytosine base of a cytosine-phosphate-guanine (CpG) dinucleotide. DNA methylation is an important epigenetic mechanism that plays a role in the regulation of gene expression, and changes in DNA methylation patterns have been associated with various diseases, including cancer.
Methylation-Specific PCR (MSP)
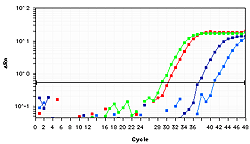
Methylation-specific PCR (MSP or MS-PCR) is a rapid and sensitive method for detecting methylation at specific CpG sites in DNA. It involves the selective methylation amplification of a target DNA fragment after bisulfite treatment (see below), using primers that are specific for the methylated form of the CpG site. MSP is a useful tool for studying DNA methylation and its potential involvement in disease.
Bisulfite Sequencing
Bisulfite sequencing is a more sensitive and comprehensive method for studying DNA methylation. It involves the conversion of unmethylated cytosines to uracils through the treatment of DNA with sodium bisulfite, while methylated cytosines are resistant to this treatment. The treated DNA is then amplified and sequenced, allowing for the determination of the methylation status of each cytosine in the sequenced region. Learn more about bisulfite sequencing.
Conclusion
Both MSP and bisulfite sequencing are important tools for understanding the role of DNA methylation in gene regulation and its potential involvement in disease. They have been widely used in research and have the potential to provide valuable insights into the underlying mechanisms of various diseases and the development of new therapeutic approaches.




 Cart (0)
Cart (0)