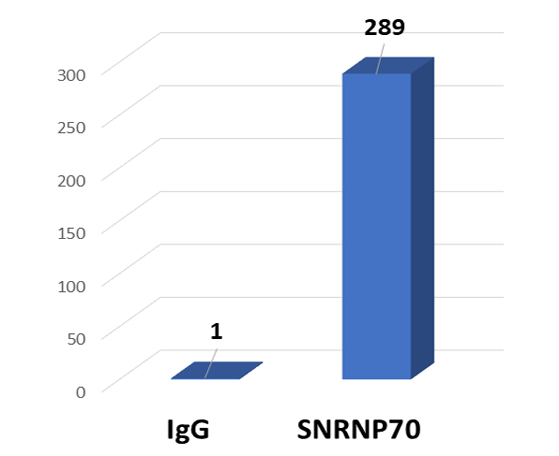
CUT&LUNCH (Cleavage Under Target and Liberate Unique Nucleic Complex Homogeneously) RNA Immunoprecipitation (RIP) is an advanced profiling technique that enables high-resolution and highly sensitive mapping of RNA-bound proteins.
The CUT&LUNCH RIP assay significantly lowers input needs, processing time, and background signal compared to conventional RIP, providing more reliable profiles.
EpigenTek delivers all-in-one CUT&LUNCH RIP and other resources to support and streamline your RNA–protein interaction studies:
- Explore the Basics: Learn the fundamentals of CUT&LUNCH RIP
- Additional Resources: Explore our detailed overview and supporting materials.
- Technical Hub: Find FAQs, troubleshooting solutions, and optimization guides in our centralized support center.




 Cart (0)
Cart (0)