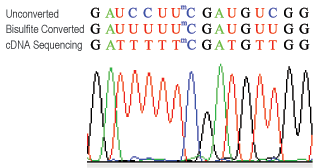
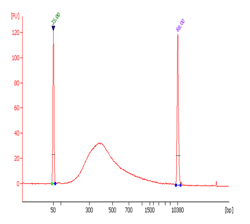
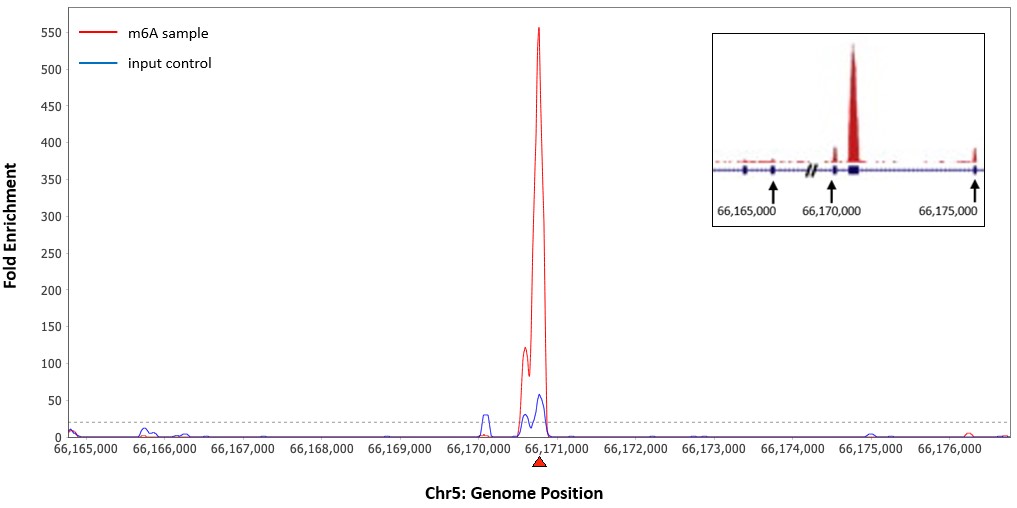
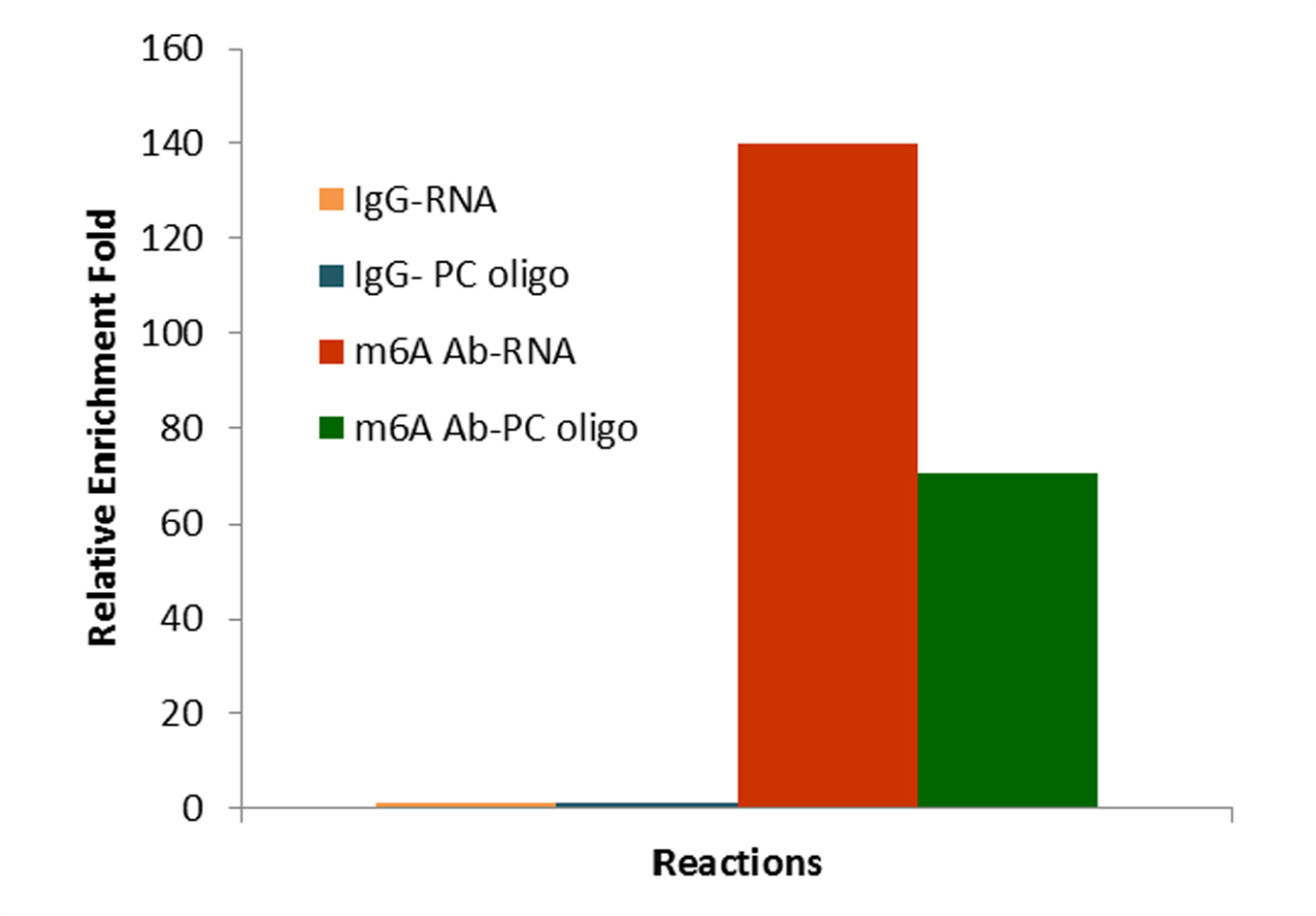
CUT&RUN m6A MeRIP (Cleavage Under Target and Recover Using Nuclease for m6A enrichment) enables rapid and highly specific enrichment of N6-methyladenosine (m6A)-containing RNA fragments for downstream analysis by PCR or next-generation sequencing.
By combining m6A immunoprecipitation with CUT&RUN-based technology, the assay achieves exceptional specificity with minimal background, even from as little as 500 ng of input RNA. Its fast, single-tube workflow minimizes sample loss and delivers enriched RNA in just ~2.5 hours with <30 minutes of hands-on time.
EpigenTek delivers all-in-one CUT&RUN m6A MeRIP and other resources to support and streamline your transcriptome-specific RNA methylation studies:
- Explore the Basics: Learn the fundamentals of CUT&RUN m6A MeRIP
- Additional Resources: Explore our detailed overview and supporting materials
- Technical Hub: Find FAQs, troubleshooting solutions, and optimization guides in our centralized support center




 Cart (0)
Cart (0)