

EpiQuik™ CUT&RUN m6A RNA Enrichment (MeRIP) Kit & EpiNext™ CUT&RUN RNA m6A-Seq Kit
Reliable, streamlined solutions for m6A enrichment and sequencing—faster workflows, higher specificity, and consistent results with minimal input.
Comprehensive m6A RNA Analysis: Enrichment and Sequencing Kits
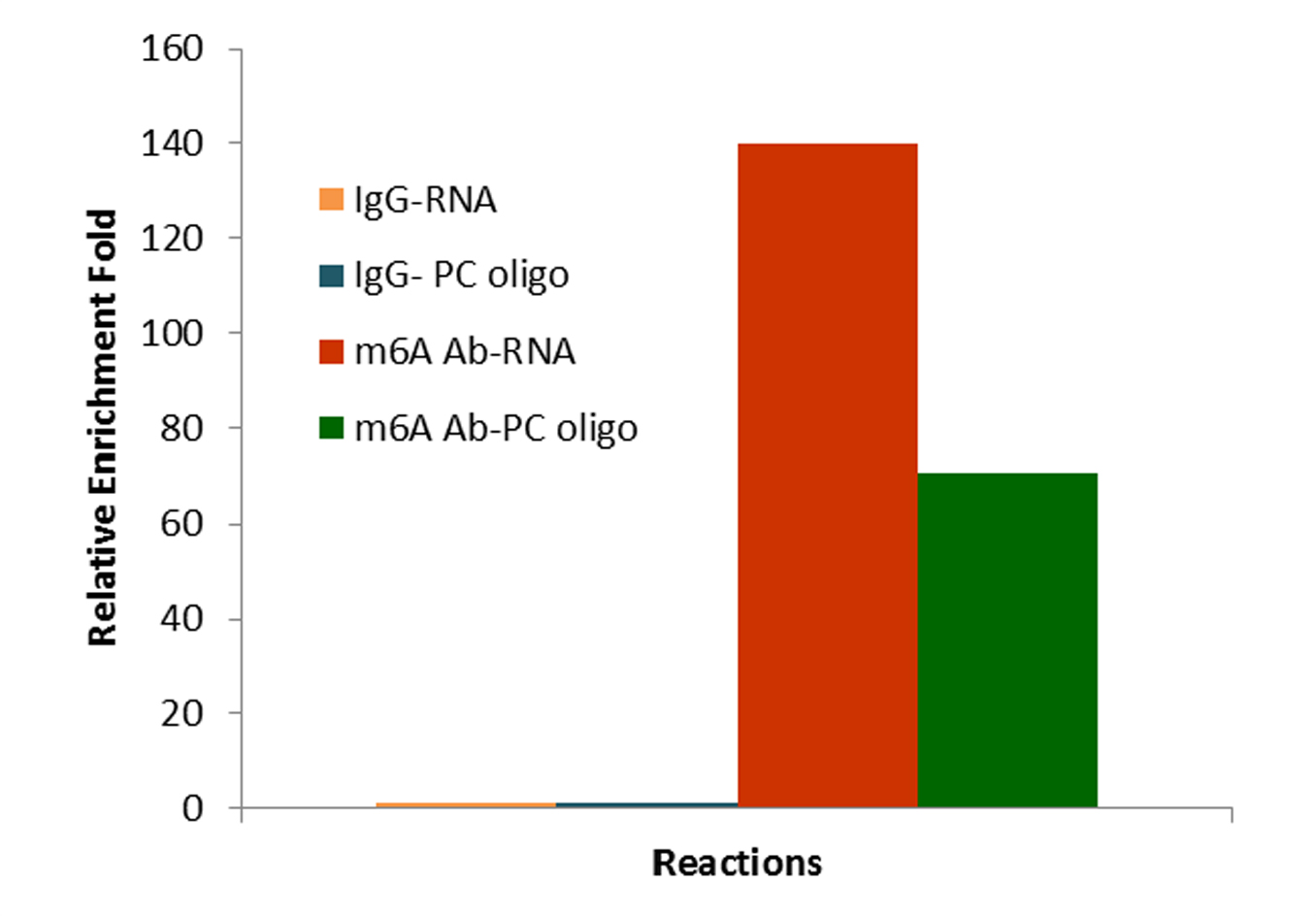
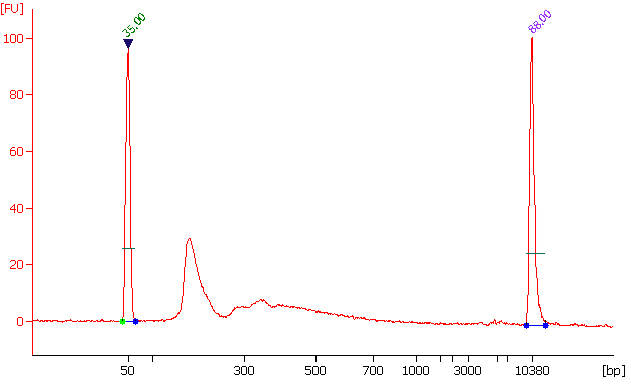
The CUT&RUN m6A RNA kits provide a next-generation approach for precise RNA methylation profiling, delivering fast, reliable, and high-resolution enrichment with minimal input.
- High enrichment: Unique cleavage enzyme mix generates short antibody-bound fragments close to m6A sites for precise mapping.
- Low input: Single-tube cleavage & capture protects targets and minimizes loss; works from as little as 500 ng RNA.
- Minimal background: Cleavage of unbound RNA at both ends reduces MeRIP-seq background—reliable analysis with <10 M reads.
- Fast & streamlined: From RNA to enriched m6A RNA in 2.5 hours (<30 min hands-on) or to library cDNA in <6 hours (<1 h hands-on).
- Highly convenient: Includes all components for cleavage, capture and (when applicable) library prep for consistent, reproducible results.
- Proven & trusted: Highly cited in over 100 peer-reviewed studies.
Consistent, convenient, and streamlined—accurate m6A profiling in hours for qRT-PCR or NGS.
Featured in the world’s leading scientific journals
Our CUT&RUN m6A RNA Enrichment (MeRIP) Kit & CUT&RUN RNA m6A-Seq Kit are trusted by leading researchers and highly cited in top-tier publications with high impact factors, fueling groundbreaking discoveries.

Impact Factor: 17.7
Shiraishi C et al. (Apr 2023). RPL3L-containing ribosomes determine translation elongation dynamics required for cardiac function. Nat Commun.
EpiNext™ CUT&RUN RNA m6A-Seq Kit
Impact Factor: 16.9
Liang Z et al. (Aug 2024). ALKBH5 governs human endoderm fate via DKK1/4-mediated Wnt/β-catenin activation. Nucleic Acids Res.
EpiQuik™ CUT&RUN MeRIP Kit
Impact Factor: 14.0
Tong Y et al. (Apr 2025). METTL3 promotes an immunosuppressive microenvironment in bladder cancer. J Immunother Cancer.
EpiQuik™ CUT&RUN MeRIP Kit
Impact Factor: 13.45
Gao X et al. (Apr 2025). MSC-derived exosomes alleviate oxidative stress-induced lysosomal membrane permeabilization damage in degenerated nucleus pulposus cells via promoting m6A demethylation of Nrf2. Free Radic Biol Med.
EpiQuik™ CUT&RUN MeRIP KitGet fast, high-specificity m6A enrichment
Explore the KitsFrequently asked questions
How much RNA input do I need?
As little as 500 ng of total RNA is sufficient for enrichment.
How long does the workflow take?
~2.5 hours for enrichment (<30 min hands-on); library cDNA in <6 hours (<1 h hands-on).
What read depth is recommended?
Reliable analysis can be achieved with fewer than 10 million reads owing to low background.
Which downstream applications are supported?
Enriched RNA is suitable for qRT-PCR and RNA-seq; the m6A-Seq kit produces sequencing-ready libraries.




 Cart (0)
Cart (0)







