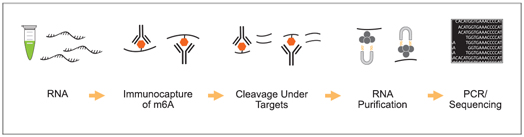
RNA modifications can occur at various locations along an RNA sequence. Identifying where they are and what impact they may have on post-transcriptional regulation, including gene expression and mRNA control, can help researchers understand anomalies in various biological functions. Therefore, techniques that can quickly and accurately detect these modifications, especially m6A in RNA, are important for epigenetic research. meRIP and MeRIP-seq are two familiar methods used to detect m6A RNA methylation in the entire transcriptome range, but they have limitations and can be pricey. EpigenTek has improved upon these techniques, developing a newer method called CUT&RUN RNA m6A enrichment (meRIP). Using a specialized nuclease that enriches for m6A-containing RNA, the process allows for high-resolution mapping in only a few hours.
What is CUT&RUN m6A meRIP?
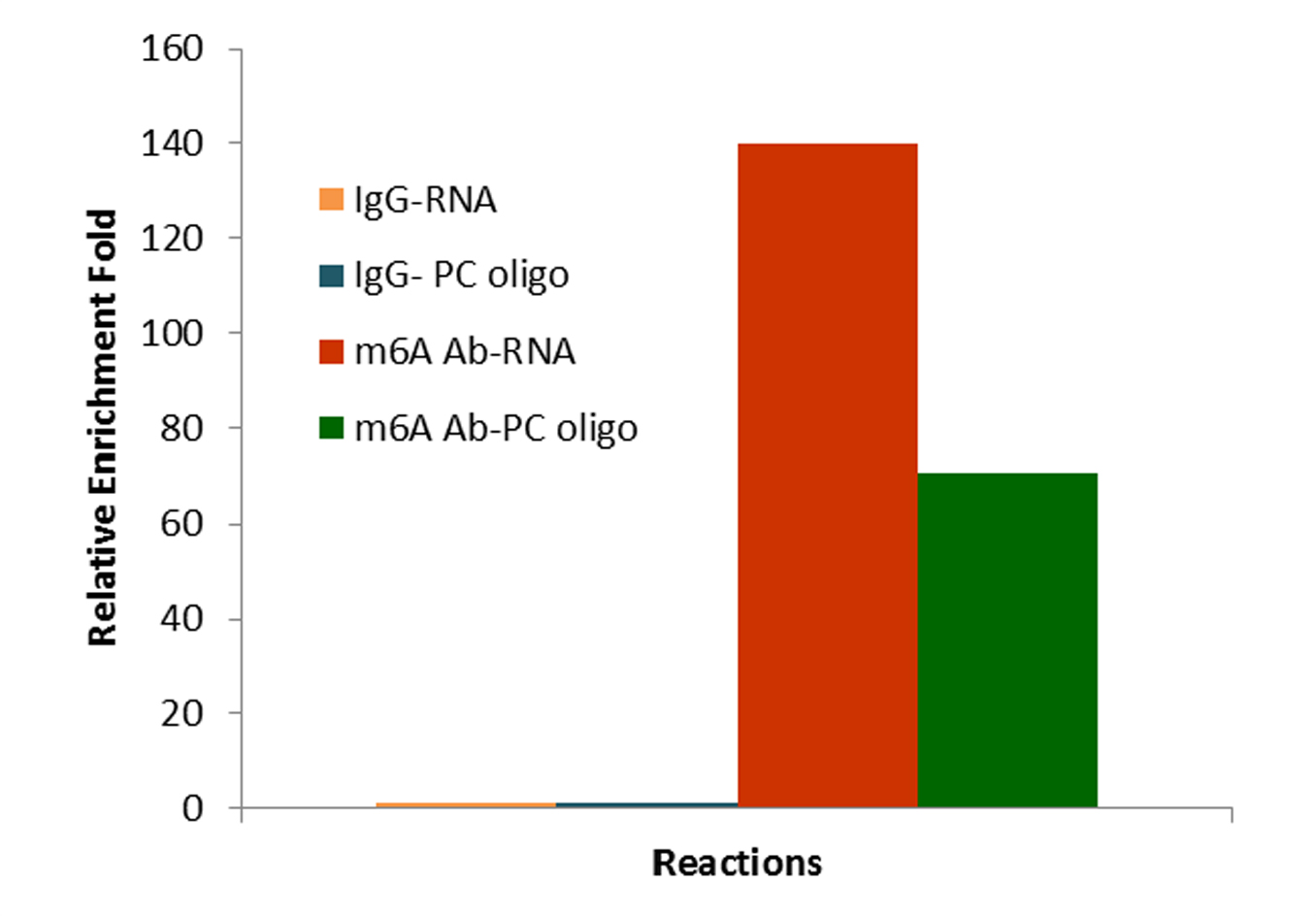
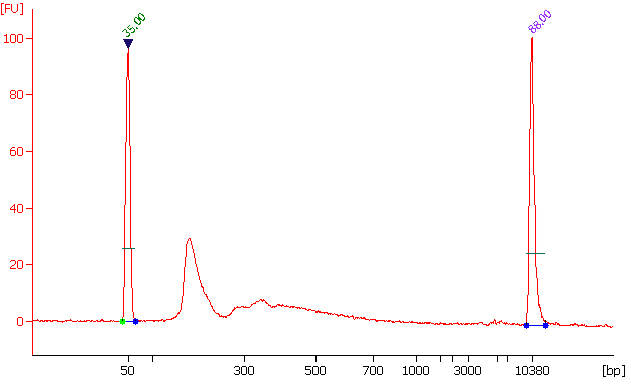
This innovative approach combines the advantages of MeRIP-seq, miCLIP, and CUT&RUN with EpigenTek’s proprietary EpiQuik technology for higher enrichment, lower input, reduced background, and a faster, more streamlined procedure. CUT&RUN m6A MeRIP uses a state-of-the-art RNA cleavage enzyme mix to simultaneously fragment immunocaptured RNA and cleave/remove any RNA sequences at both ends of the target m6A-containing sequences without affecting RNA regions occupied by the antibody. Short RNA fragments are consequently generated only bound with anti-m6A antibody. True target m6A-enriched regions can therefore be reliably identified, and high-resolution mapping achieved.
This innovative approach combines the advantages of MeRIP-seq, miCLIP, and CUT&RUN with EpigenTek’s proprietary EpiQuik technology for higher enrichment, lower input, reduced background, and a faster, more streamlined procedure. CUT&RUN m6A MeRIP uses a state-of-the-art RNA cleavage enzyme mix to simultaneously fragment immunocaptured RNA and cleave/remove any RNA sequences at both ends of the target m6A-containing sequences without affecting RNA regions occupied by the antibody. Short RNA fragments are consequently generated only bound with anti-m6A antibody. True target m6A-enriched regions can therefore be reliably identified, and high-resolution mapping achieved.
Featured Kits
EpiQuik CUT&RUN m6A RNA Enrichment (MeRIP) Kit
High resolution m6A RNA enrichment based on CUT&RUN for increased specificity and signals
- Fast: Just 2.5 hours
- Low Sample Input: As low as 500 ng
- High Resolution/ Low Background: Target m6A RNA cleaved at both ends
- Convenient: All-in-one kit for enriching m6A RNA
EpiNext CUT&RUN RNA m6A-Seq Kit
All-in-one m6A RNA enrichment and NGS library preparation based on CUT&RUN
- Fast: Less than 6 hours
- Low Sample Input: As low as 500 ng
- High Resolution/ Low Background: Target m6A RNA cleaved at both ends
- Convenient: m6A RNA capture + NGS library prep
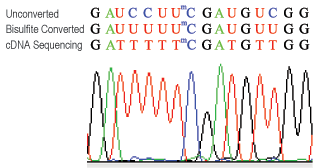
Methylamp RNA Bisulfite Conversion Kit
For robust, chemical-based bisulfite treatment of RNA for methylated cytosine analysis
- Fast: Only 3 hours
- High Conversion Efficiency: >99.9%
- Low Sample Input: As low as 5 ng
- Low Degradation: Prevents >90% RNA loss




 Cart (0)
Cart (0)