DNA methylation is an epigenetic modification where a methyl group is added to cytosine bases, typically at CpG sites, by DNA methyltransferases (DNMTs), influencing gene expression without altering the DNA sequence. This modification can be dynamically regulated, as ten-eleven translocation (TET) enzymes catalyze the oxidation of 5-methylcytosine (5-mC) into hydroxymethylcytosine (5-hmC) and further into formylcytosine (5-fC) and carboxylcytosine (5-caC), facilitating active DNA demethylation.
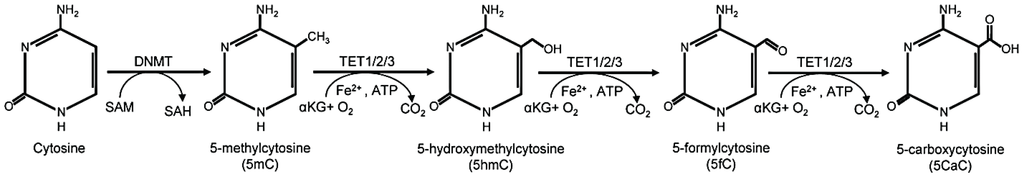
Chemical pathway illustrating cytosine methylation by DNMT and demethylation by TET enzymes, showing 5mC, 5hmC, 5fC, and 5caC.
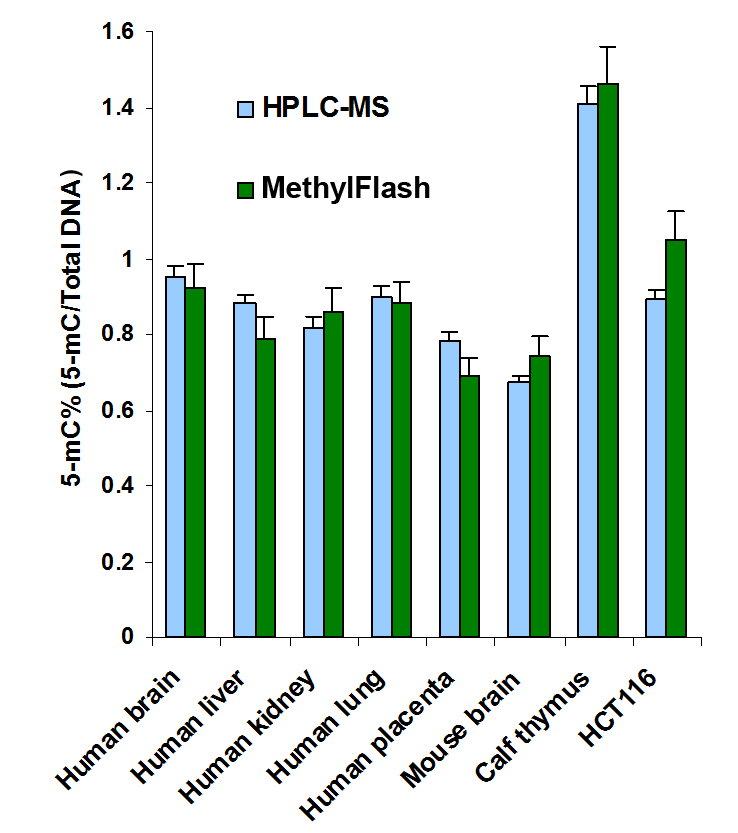
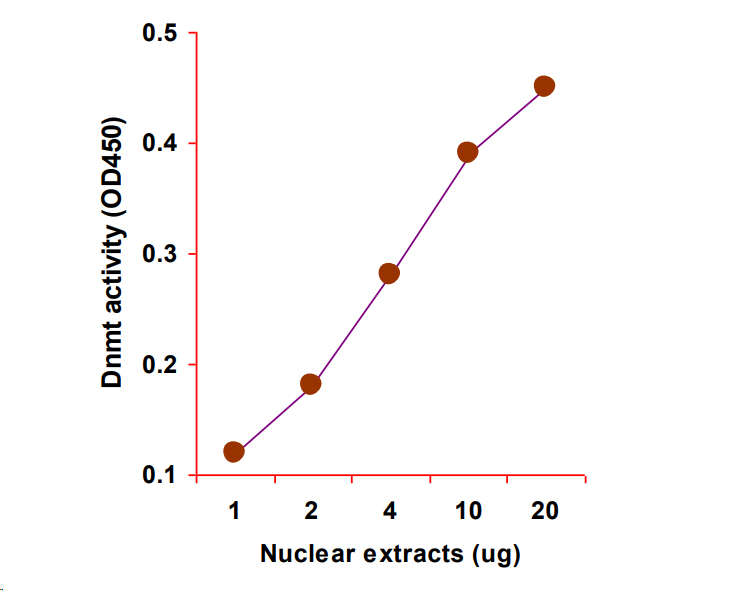
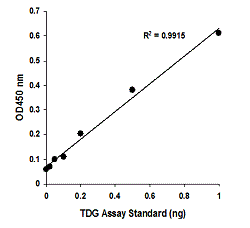
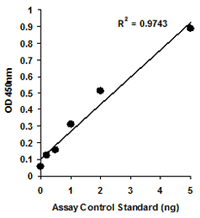
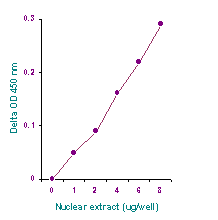
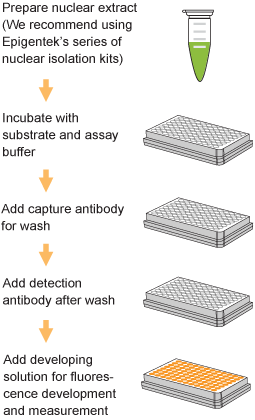
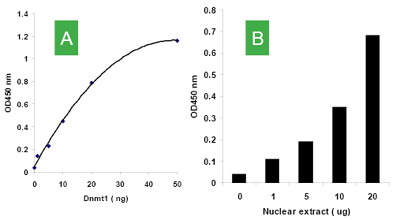
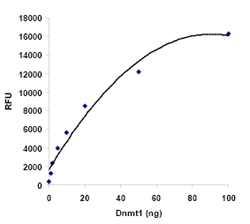
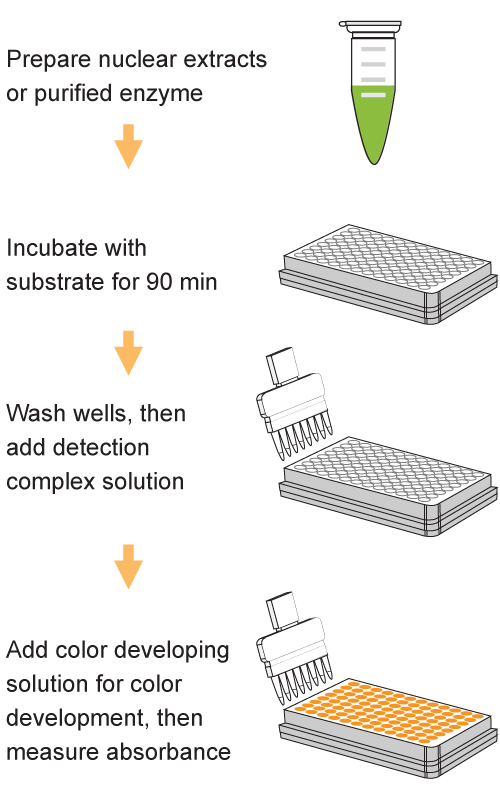
Quantify levels of 5-mC, 5-hmC, 5-fC, or 8-OHdG in total DNA
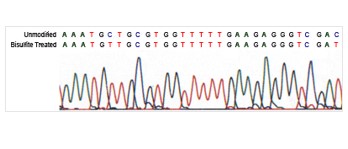
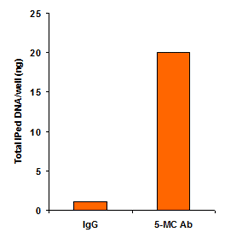
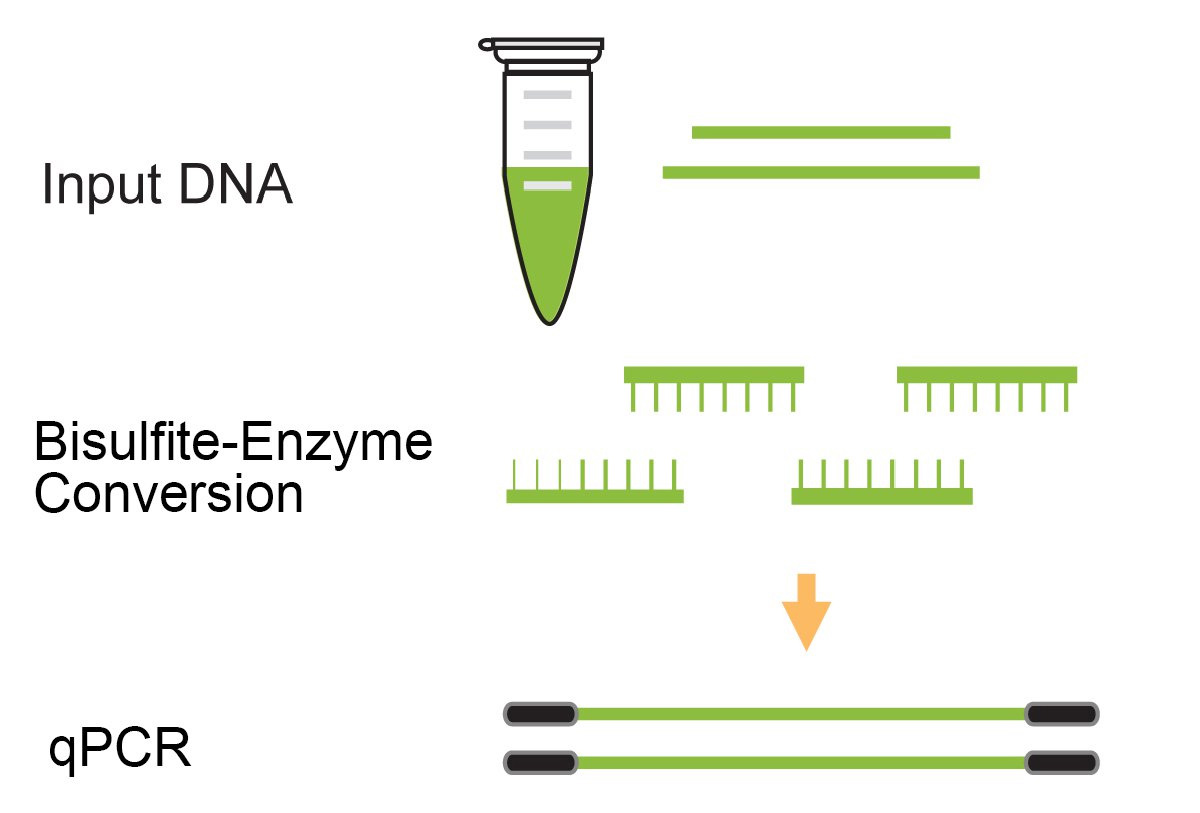
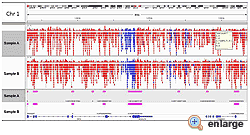
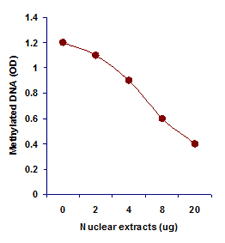
Differentiate between methylated and unmethylated cytosines gene specifically or genome wide




 Cart (0)
Cart (0)