Radioactive contamination in the world’s oceans remains a growing environmental concern, particularly near nuclear facilities and waste discharge sites. Among these pollutants, tritium—a radioactive form of hydrogen produced during nuclear energy generation—has drawn attention for its ability to enter marine ecosystems in both dissolved and particle-bound forms. While tritium’s radioactive effects are well known, its subtler biological consequences at the molecular level have been less explored.
In a study published in Environmental Pollution, researchers investigated how exposure to tritiated water (HTO) and tritium-bound stainless-steel particles (T-SSPs) affects marine mussels (Mytilus galloprovincialis). Their goal was to understand both genotoxic effects (DNA damage) and epigenetic changes, focusing on global DNA methylation, a key indicator of how environmental stress can alter gene regulation.
Measuring Global DNA Methylation
To quantify changes in global methylation, the researchers used the MethylFlash™ Global DNA Methylation (5-mC) ELISA Easy Kit from EpigenTek. This high-sensitivity, ELISA-style platform enabled accurate measurement of 5-methylcytosine (% 5-mC) in mussel gill and digestive gland tissues, allowing for precise comparison between control and tritium-exposed groups.
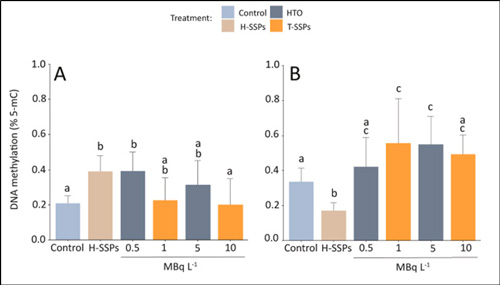
Following exposure, the mussels exhibited both increased DNA damage and elevated global DNA methylation, particularly in gill tissues. This analysis highlighted global DNA methylation as a sensitive biomarker for detecting the molecular impact of environmental radiation.
Key Findings
The study revealed a clear link between radioactive exposure and altered global DNA methylation in marine mussels. After seven days of tritium exposure, mussels showed significant increases in global 5-mC levels, particularly in the gills, along with dose-dependent changes that indicated greater epigenetic disruption at higher concentrations. These methylation shifts occurred alongside measurable DNA damage, suggesting that tritium not only harms DNA directly but also influences how genes are regulated in response to radiation-induced stress. Together, these findings demonstrate that monitoring global DNA methylation provides valuable insight into how environmental contaminants affect gene regulation and organismal health at the molecular level.
Future Outlook
Building on these findings, this research underscores the importance of incorporating epigenetic biomarkers like DNA methylation into environmental monitoring programs. Future studies may explore whether these methylation changes are reversible, persist across generations, or contribute to long-term ecological effects. Expanding such analyses across species could help scientists better assess the global impact of radioactive and chemical pollution on marine life.
Advancing Epigenetic Discovery with EpigenTek
The MethylFlash™ Global DNA Methylation (5-mC) ELISA Easy Kit simplifies and accelerates global methylation analysis—delivering publication-ready results in just hours. Its high sensitivity, strong reproducibility, and streamlined workflow make it an essential tool for researchers studying how environmental, developmental, or disease-related factors influence the epigenome.
EpigenTek offers a complete portfolio of global DNA methylation solutions, including the MethylFlash™ Global DNA Hydroxymethylation (5-hmC) ELISA Easy Kit, as well as assays for oxidative stress damage (as demonstrated in this study). Together, these products empower scientists to explore the full spectrum of epigenetic modifications with unmatched precision, speed, and reliability.




 Cart (0)
Cart (0)