Understanding how proteins interact with chromatin is central to epigenetics research, but traditional methods like ChIP-seq often require large numbers of cells, lengthy workflows, and extensive optimization. Enter CUT&LUNCH (Cleavage Under Target and Liberate Unique Nucleic Complex Homogeneously) — a cutting-edge approach that streamlines chromatin profiling while maintaining high specificity and sensitivity.
What is CUT&LUNCH?
CUT&LUNCH is a next-generation chromatin profiling method that leverages a unique nucleic acid cleavage enzyme mix to selectively fragment DNA at both ends of target protein/DNA interaction regions. Cells are permeabilized and incubated with a ChIP-grade antibody targeting a protein of interest. Cleaved and unbound DNA is then removed, leaving only the antibody-bound complexes intact. This selective approach significantly reduces background noise and enables researchers to work with very low cell numbers.
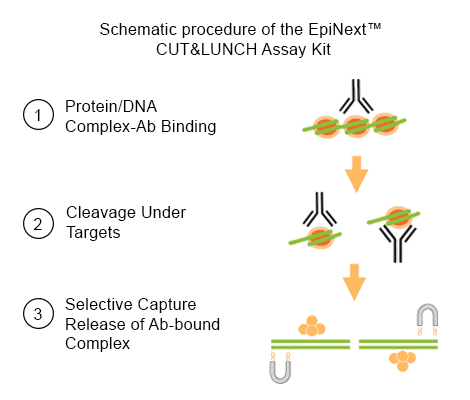
Unlike traditional ChIP, CUT&LUNCH does not require bead immobilization or crosslinking, and unlike CUT&RUN or CUT&Tag, it minimizes non-specific cleavage and sequence bias, resulting in cleaner, more reproducible data.
Applications and Advantages
• Low-input profiling: CUT&LUNCH can generate meaningful results from as few as 2,000 cells, making it ideal for rare or primary cell populations.
• Rapid workflow: The assay can be completed in under 2 hours, and the full CUT&LUNCH-Seq workflow with library prep can be done in less than 3 hours, enabling same-day results.
• Flexible downstream analysis: DNA fragments recovered can be analyzed via qPCR for targeted studies or prepared into sequencing-ready libraries for genome-wide mapping.
• High specificity and low background: By removing unbound DNA and selectively recovering only antibody-bound complexes, CUT&LUNCH provides enhanced signal-to-noise ratios, which is particularly valuable for detecting low-abundance proteins.
CUT&LUNCH in Practice
CUT&LUNCH has been successfully applied to profile histone modifications such as H3K4me3 and H3K9me3, as well as transcription factors like RNA Polymerase II, with results consistent with conventional ChIP-seq but using far fewer cells and a fraction of the time. This efficiency opens doors for experiments previously limited by sample availability or workflow complexity.
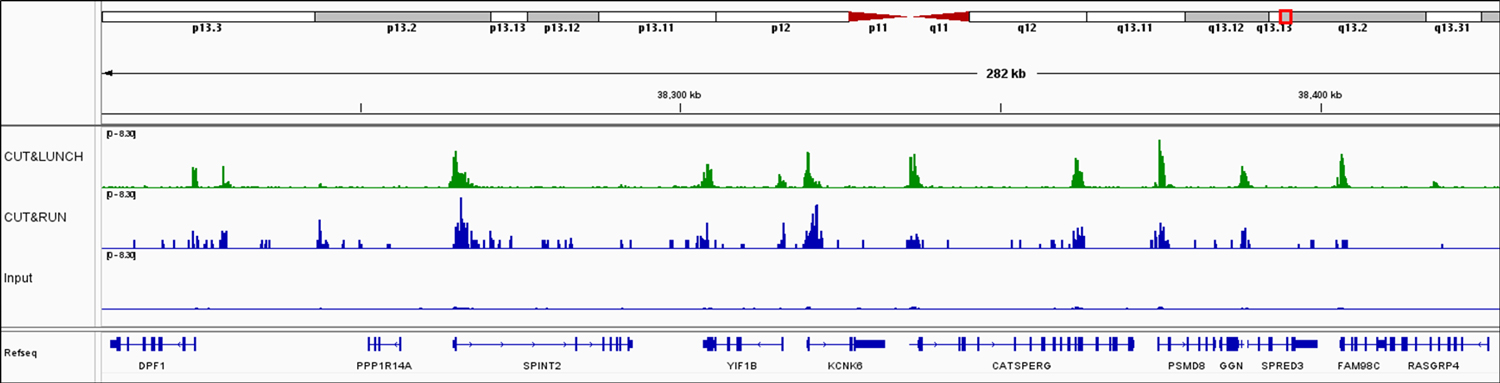
CUT&LUNCH outperforms CUT&RUN. Peak scores are plotted in the bar graph for 100,000 MCF-7 cells using CUT&LUNCH, followed by NGS, for H3K4me3. The traditional CUT&RUN assay was used as a comparison.
CUT&LUNCH TF (RNA Polymerase II) peak profiles are consistent with ENCODE ChIP-seq profiles. CUT&LUNCH: Peak scores are plotted in the bar graph for 0.1 million MCF-7 cells. ENCODE ChIP-Seq: Peak scores are plotted in the bar graph for 5 million cells.
Whether you are studying epigenetic regulation, or transcription factor binding, CUT&LUNCH offers a streamlined, sensitive, and reproducible alternative to traditional chromatin profiling methods.
Learn more:
• EpiNext™ CUT&LUNCH-Seq Kit (P-2033)




 Cart (0)
Cart (0)













