Oga K et. al. (June 2024). Involvement of dysregulated hippocampal histone H3K9 methylation at the promoter of the BDNF gene in impaired memory extinction Psychopharmacology (Berl).
This study explores how histone H3 lysine 9 dimethylation (H3K9me2) affects brain-derived neurotrophic factor (BDNF) expression in the hippocampus, contributing to impaired extinction of fear memory (EFM) in PTSD. Using a PTSD rat model, researchers found decreased BDNF mRNA levels and increased H3K9me2 at BDNF promoter IV after extinction training. The histone methyltransferase inhibitor BIX01294 restored BDNF mRNA levels and improved EFM in these rats. The results indicate that dysregulated H3K9me2 at the BDNF promoter plays a role in PTSD, and BIX01294 could be a potential treatment.
Products Used: EpiQuik Total Histone Extraction Kit, EpiQuik Chromatin Immunoprecipitation (ChIP) Kit
Xin X et. al. (June 2024). Temperature-dependent jumonji demethylase modulates flowering time by targeting H3K36me2/3 in Brassica rapa Nat Commun. 15(1):5470.
This study explores how global warming affects flowering time and yield in Brassica rapa. The gene BrJMJ18, encoding an H3K36me2/3 demethylase, was identified as crucial for thermotolerance. The allele from thermotolerant Caixin (BrJMJ18Par) influences flowering and growth based on temperature, modulates BrFLC3 expression, and affects chlorophyll biosynthesis under heat stress. This suggests BrJMJ18 enhances heat resilience in Brassica rapa without impacting normal agronomic traits.
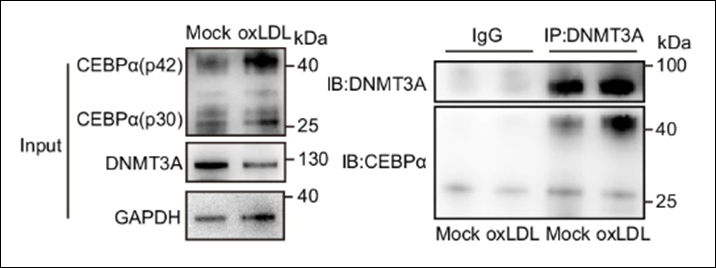
Seehawer M et. al. (June 2024). Loss of Kmt2c or Kmt2d drives brain metastasis via KDM6A-dependent upregulation of MMP3 Nat Cell Biol.
This study examines the role of KMT2C and KMT2D, genes encoding histone H3 lysine 4 methyltransferases, in triple-negative breast cancer (TNBC) metastasis. Deletion of Kmt2c or Kmt2d in non-metastatic TNBC mouse models led to increased brain metastasis. Chromatin profiling and sequencing revealed changes in H3K4me1, H3K27ac, and H3K27me3 marks, with increased binding of the H3K27me3 demethylase KDM6A, correlating with gene expression changes. Mmp3 was identified as a gene commonly upregulated through these epigenetic changes. Inhibition of KDM6A or Mmp3 reduced metastasis, highlighting the KDM6A-MMP3 axis as a key driver of KMT2C/D loss-driven metastasis in TNBC.
Products Used: EpiQuik Total Histone Extraction Kit
Flores-Sierra JJ et. al. (June 2024). The DNA methyltransferase inhibitor decitabine blunts the response to a high animal fat and protein diet in mice J Lipid Res. :100586.
This study investigates the impact of the DNA methyltransferase inhibitor decitabine (Dec) on mice fed a high-animal fat and protein diet (HAFPD), a model for Western-type diets associated with cardiovascular risk. Over eight weeks, Dec significantly reduced body weight gain in female mice and affected lipid distribution by increasing muscle lipid content while decreasing liver fat. It also improved mitochondrial function but caused adipose insulin resistance. The effects of Dec varied based on tissue type and exposure duration. These findings suggest Dec might redirect lipid accumulation to skeletal muscle due to enhanced mitochondrial function, offering potential insights for treating lipid dysmetabolism.
Products Used: MethylFlash Methylated DNA 5-mC Quantification Kit (Colorimetric)
Zhou Y et. al. (June 2024). CELLULOSE SYNTHASE-LIKE C proteins modulate cell wall establishment during ethylene-mediated root growth inhibition in rice Plant Cell.
This study shows that ethylene regulates cell wall establishment in rice roots by inducing cell wall thickening and upregulating CELLULOSE SYNTHASE-LIKE C (OsCSLC) and CELLULOSE SYNTHASE A (OsCESA) genes. OsCSLC2 and its homologs are crucial for ethylene-mediated xyloglucan biosynthesis, which restricts cell wall extension. OsCSLC2 functions downstream of ETHYLENE-INSENSITIVE3-LIKE1 (OsEIL1), which activates OsCSLC genes. The auxin signaling pathway also modulates these processes, linking hormone signaling with cell wall formation and enhancing our understanding of root growth in rice.
Products Used: EpiQuik Plant ChIP Kit
Yan Z et. al. (June 2024). YTHDC2 mediated RNA m6A modification contributes to PM2.5-induced hepatic steatosis Yan Z et. al. (June 2024)
In this study, researchers investigated how exposure to PM2.5 during the heating season contributes to hepatic steatosis. They found that PM2.5 exposure increased m6A methylation levels in liver cells, leading to lipid accumulation. This effect was associated with reduced levels of YTHDC2, a protein that normally stabilizes mRNA of lipid-related genes like CEPT1 and YWHAH. Restoring YTHDC2 reversed these effects, highlighting its role in protecting against PM2.5-induced liver fat accumulation.
Products Used: EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric)
Chen Q et. al. (June 2024). WTAP/YTHDF1-mediated m6A modification amplifies IFN-γ-induced immunosuppressive properties of human MSCs J Adv Res.
This study investigates how interferon-γ (IFN-γ) amplifies the immunosuppressive capabilities of mesenchymal stem cells (MSCs) through N6-methyladenosine (m6A) modification. Researchers examined the role of WTAP in regulating m6A levels in MSCs licensed by IFN-γ. They found that IFN-γ induces m6A methylation on genes like IDO1, PD-L1, ICAM1, and VCAM1, crucial for MSC-mediated immunosuppression. WTAP deficiency impairs this process, reducing MSCs' ability to suppress activated T cells. Furthermore, enhancing WTAP/YTHDF1 expression enhances the therapeutic efficacy of IFN-γ-licensed MSCs in colitis and arthritis models, suggesting potential advancements in MSC-based therapies through epitranscriptomic regulation.
Products Used: EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric)
Le M et. al. (June 2024). m6A-YTHDF1 Mediated Regulation of GRIN2D in Bladder Cancer Progression and Aerobic Glycolysis Biochem Genet.
This study explores how m6A modification, guided by YTHDF1, regulates GRIN2D in bladder cancer. Researchers found increased m6A levels in bladder cancer tissues and showed that YTHDF1 positively influences GRIN2D expression. Functionally, GRIN2D promotes cancer cell growth and enhances aerobic glycolysis. Inhibiting the m6A-YTHDF1-GRIN2D pathway suppressed cancer progression and metabolic changes. These findings highlight the pathway's role in bladder cancer biology and suggest potential therapeutic targets for future treatments.
Products Used: EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric)
Fan S et. al. (July 2024). METTL14-mediated Bim mRNA m6A modification augments osimertinib sensitivity in EGFR-mutant NSCLC cells Mol Cancer Res.
This study investigates how m6A modification of Bim mRNA, mediated by METTL14, affects osimertinib resistance in EGFR-mutant NSCLC. Researchers found that reduced METTL14 levels contribute to osimertinib resistance, while restoring METTL14 enhances drug sensitivity by stabilizing Bim mRNA and promoting apoptosis. Clinical findings support METTL14's role, showing its decreased expression in osimertinib-resistant NSCLC tissues correlates with poor prognosis. These insights suggest potential therapeutic avenues to combat resistance to EGFR-targeted therapies like osimertinib.
Products Used: EpiQuik m6A RNA Methylation Quantification Kit (Colorimetric)
Xu R et. al. (June 2024). Development of a targeted method for DNA adductome and its application as sensitive biomarkers of ambient air pollution exposure J Hazard Mater. 476:135018.
This study introduces a precise method for analyzing DNA adducts, crucial markers of environmental carcinogen exposure. Using advanced UHPLC-QqQ-MS/MS technology, researchers developed a robust approach to detect 41 DNA adducts with high accuracy and sensitivity. Application of this method to samples from pregnant women in Shanxi and Beijing identified up to 23 DNA adducts, revealing potential impacts of ambient air pollution on DNA methylation and oxidative damage. These findings underscore DNA adducts as sensitive biomarkers for assessing air pollution exposure in epidemiological research.
Products Used: EpiQuik One-Step DNA Hydrolysis Kit




 Cart (0)
Cart (0)