Myeloid-derived suppressor cells (MDSCs) have long been recognized as key players in cancer progression. These cells, which usually inhibit immune responses and promote tumor growth, are a central focus of cancer research. However, the exact mechanisms underlying their function, particularly in colorectal cancer (CRC), remain somewhat elusive. Recent studies have begun to shed light on how specific molecular modifications might influence MDSC behavior. One such modification involves N6-methyladenosine (m6A), a prevalent RNA modification with significant implications for gene expression and cancer biology.

The m6A modification is an epigenetic mark added to RNA by methyltransferases, removed by demethylases, and recognized by various binding proteins, collectively known as "writers," "erasers," and "readers." ALKBH5, a member of the "eraser" group, is particularly intriguing. This demethylase removes m6A from RNA molecules, influencing their stability and function. Dysregulation of ALKBH5 has been linked to several cancers, including breast, pancreatic, and gastric cancer. However, its role in MDSCs, particularly in the context of CRC, has not been thoroughly explored until now.
A study published in Non-coding RNA Research explored the impact of ALKBH5 on MDSCs in CRC. The researchers discovered that MDSCs in CRC mouse models displayed increased m6A modification levels and reduced ALKBH5 expression. Overexpressing ALKBH5 in these cells led to a notable reduction in their immunosuppressive capabilities and a decrease in their protumorigenic activities.
Using the EpiQuik™ CUT&RUN m6A RNA Enrichment (MeRIP) Kit, the study revealed that ALKBH5 overexpression significantly reduced m6A modification on Arg-1 mRNA. This decrease in m6A levels resulted in diminished mRNA stability and lower Arg-1 protein levels, which in turn reduced the enzyme’s ability to suppress T cell activity, ultimately impairing tumor growth.
Further validation with the EpiQuik™ m6A RNA Methylation Quantification Kit showed that mice with ALKBH5-overexpressing MDSCs had reduced tumor growth compared to control groups. This suggests that ALKBH5 not only modulates MDSC function but also represents a promising target for therapeutic intervention.
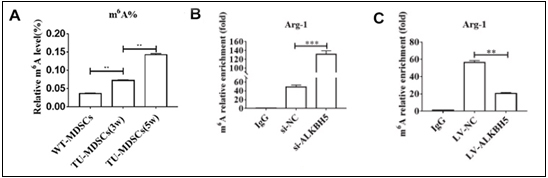
The study revealed that ALKBH5’s impact on MDSCs is mediated through its effect on Arg-1. By demethylating m6A modifications on Arg-1 mRNA, ALKBH5 reduces the stability and expression of this key immunosuppressive factor. This action weakens the MDSCs’ ability to inhibit T-cell activity and promote tumor growth.
These findings open new avenues for cancer treatment by targeting MDSCs and modulating ALKBH5 levels. The study highlights ALKBH5 as a potential therapeutic target, offering a fresh perspective on the molecular mechanisms of MDSC function and advancing our understanding of CRC immunology. By elucidating the role of RNA modifications in immune cell regulation, this research provides promising new strategies for enhancing immunotherapy and reducing tumor progression.




 Cart (0)
Cart (0)