Bisulfite Conversion & Methylated DNA Immunoprecipitation
Epigenetics, the study of heritable changes in gene expression that do not alter DNA sequence, has uncovered complex regulatory layers governing gene function. Among the various mechanisms, DNA methylation—most commonly at cytosine residues in CpG dinucleotides—serves as a critical epigenetic mark influencing gene expression and chromatin structure. DNA methylation patterns are extremely dynamic, modifiable in response to environmental factors, and implicated in numerous biological pathways and diseases, including cancer, neurodegenerative disorders, and developmental abnormalities. Given the significant role of methylation in gene regulation, studying these modifications at high resolution has become essential for advancing our understanding of gene function and epigenetic memory.
Principles and Applications of Bisulfite Conversion and Immunoprecipitation in Methylation Analysis
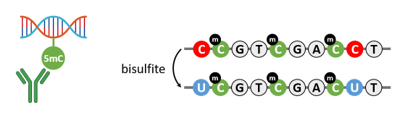
Bisulfite conversion is a cornerstone technique in DNA methylation analysis that provides single-base resolution of methylated cytosines. During this process, unmethylated cytosines are chemically converted to uracils, while 5-methylcytosine (5-mC) remains unaffected. Subsequent to conversion, downstream applications such as PCR or NGS can reveal methylation arrangements at specific genomic loci, providing a detailed landscape of methylation status.
In addition to 5-mC, the hydroxymethylated form of cytosine (5-hmC) has gained attention as an intermediate in active DNA demethylation and a marker for various cellular states. MeDIP (methylated DNA immunoprecipitation) and hMeDIP (hydroxymethylated DNA immunoprecipitation) assays leverage antibodies specific to 5-mC and 5-hmC, respectively, to enrich methylated regions of the genome. This immunoprecipitation-based approach, when followed by sequencing or quantitative PCR, enables targeted analysis of methylation across the genome, particularly useful for site-specific studies or for discovering differentially methylated regions.
Reliable Solutions for Precise DNA Methylation Profiling
Traditional bisulfite conversion methods often suffer from DNA degradation due to the harsh chemical treatment, low conversion efficiency, and incomplete conversion, which can lead to inaccurate PCR or NGS analyses. Additionally, current MeDIP and hMeDIP procedures often face challenges in antibody specificity, inconsistent enrichment of low-abundance methylation marks, and technical variability, thus complicating quantitative analysis and comparisons between samples.
EpigenTek offers a range of solutions optimized for DNA methylation analysis. Our bisulfite conversion kits streamline the conversion process with greater efficiency and minimal DNA loss, ensuring high fidelity for downstream PCR and sequencing applications. For immunoprecipitation needs, our MeDIP and hMeDIP kits provide reliable enrichment of 5-mC and 5-hmC, empowering researchers to analyze site-specific DNA methylation patterns accurately. Each kit is optimized for sensitivity and ease of use, making EpigenTek a preferred partner in epigenetic research. Explore these kits to elevate your DNA methylation studies to the next level of precision and reliability.




 Cart (0)
Cart (0)