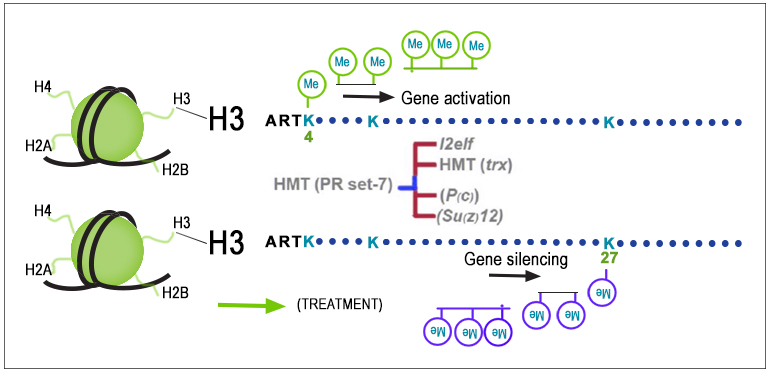
Epigenetic regulation through histone post-translational modifications (PTMs) is a cornerstone of gene expression control. The dynamic and reversible nature of these modifications, which comprise acetylation, methylation, phosphorylation, and ubiquitination, influence cellular functions and biological processes. To elucidate the profound implications of histone PTMs in health and disease, robust quality methods for their quantification are indispensable.

Applications
Histone PTM quantification is integral to basic science, helping to unravel the dynamic changes in these epigenetic marks during normal physiological activities such as embryonic development and tissue differentiation. Studying these modifications provides crucial insights into, for example, the regulatory mechanisms guiding lineage commitment and organogenesis, or the role of epigenetics in tissue-specific gene expression programs. In disease research, quantifying histone PTMs is instrumental in deciphering altered epigenetic landscapes. This approach contributes to diagnostic strategies by identifying specific modification signatures associated with different disease subtypes. Moreover, understanding the epigenetic underpinnings of diseases enables the identification of potential therapeutic targets, paving the way for personalized treatment strategies.
Quantification of histone PTMs also plays a pivotal part in drug discovery efforts, notably in screening for small molecules that modulate these epigenetic marks. Identifying compounds that influence histone PTMs offers prospective remedial avenues for various diseases, like cancer and neurological disorders. The applications extend to the progression of epigenetic-targeted therapies.
Methods
Diverse methodologies are utilized in histone PTM quantification. Chromatin Immunoprecipitation (ChIP) stands as a foundational method for this purpose, employing a selective immunoprecipitation tactic. This technique encompasses cross-linking proteins to DNA, fragmenting the chromatin, and using specific antibodies to precipitate histones bearing the modification of interest. Following reversal of the cross-links, the enriched DNA can be analyzed downstream via PCR or next-generation sequencing. ChIP allows the mapping of histone PTMs to specific genomic loci, providing insights into their spatial distribution.
Mass Spectrometry (MS) is a versatile alternative for identifying and quantifying histone PTMs. It entails the extraction of histones from cells or tissues, enzymatic digestion, and the analysis of resulting peptides through mass spectrometry. MS allows the simultaneous analysis of multiple histone PTMs, providing a comprehensive overview of the epigenetic landscape. Enzyme-Linked Immunosorbent Assay (ELISA) offers a high-throughput means to histone PTM quantification, relying on the specific binding of antibodies to modified histones. During a typical ELISA, histones from samples are immobilized onto a solid phase, followed by detection with specific antibodies. ELISA is particularly advantageous for large-scale screening of histone PTM levels in various biological samples.
Challenges
Despite certain advancements, drawbacks in histone PTM quantification methods still persist that impact the accuracy and reliability of results. One of the primary challenges is ensuring the specificity and sensitivity of antibodies. The accuracy of results depends on the selectivity of antibodies to the modification of interest. Limited availability of well-validated antibodies for certain modifications necessitates ongoing efforts in antibody characterization and validation.
Ensuring data reproducibility is another critical concern in histone PTM quantification. Standardizing protocols and analytical pipelines is essential to mitigate inter-laboratory variability and enhance the comparability of results across different studies. Background noise in experimental procedures and the need for robust normalization strategies to account for variations in experimental conditions are crucial considerations as well. Achieving precise quantification, especially in the context of dynamic cellular states, requires the development of standardized normalization practices.
To address these issues, EpigenTek has designed a series of highly sensitive, specific, reliable, and consistent kits for histone PTM quantification. With the Histone H3 Modification Multiplex Assay Kit and the Histone H4 Modification Multiplex Assay Kit , either 21 different histone H3 PTMs or 10 different histone H4 PTMs, which include all of the most important and well-characterized patterns, can be simultaneously screened and measured on the same microplate using a quick and efficient ELISA-like format. These innovative colorimetric assays feature a high sensitivity with a low detection threshold, and, in addition, preclude the need for electrophoresis, chromatography, or expensive equipment. For more in-depth analysis of a specific histone mark, singleplex kits are also available for each of the modifications included in the multiplex panels.



 Cart (0)
Cart (0)