DNA methylation and RNA methylation are extensively studied epigenetic modifications with well-established gene regulatory functions. These modifications are essential for maintaining cellular homeostasis and biological health by modulating gene expression, ensuring genomic integrity, and orchestrating developmental processes. Consequently, perturbations in the homeostatic states of these marks underlie many human diseases, making DNA and RNA methylation of great medical value and focal points for drug development.

Tumorigenesis and Epigenetic Changes
Tumorigenesis is typically characterized by an overall decrease in DNA methylation, particularly 5-methylcytosine (5mC), with abnormally hypermethylated levels in the CpG island regions of specified genes. The hypomethylation observed in proto-oncogenes and the increase in DNA methylation exhibited by tumor suppressor genes, respectively enhancing and hindering the expression of these genes during cancer formation, conjointly promote tumor growth. The detection of these anomalous methylation patterns in bodily fluids (liquid biopsies) has found clinical utility as diagnostic and prognostic biomarkers.
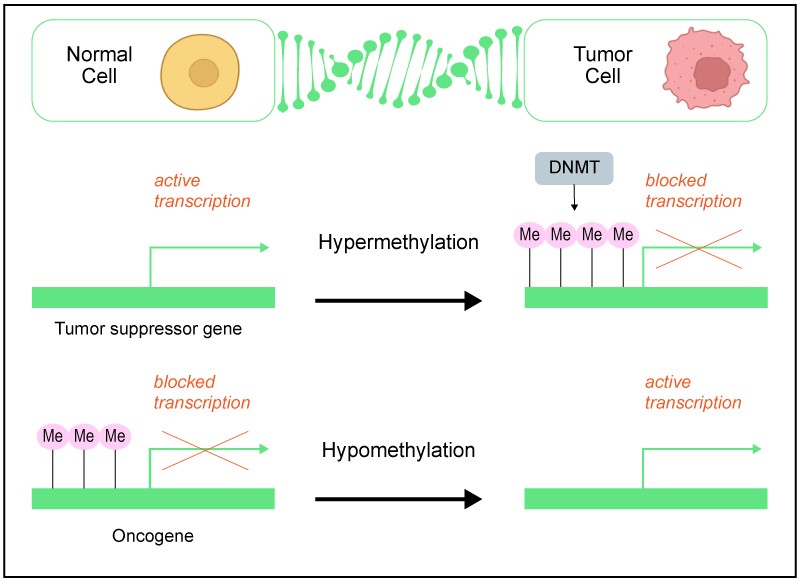
Therapeutic Targeting of DNA Methylation
Therapeutic targeting of the DNA methylation pathway has also garnered healthcare focus. DNA methyltransferase inhibitors (DNMTis) were one of the earliest epigenetic drugs used in cancer treatment. Azacytidine and decitabine are FDA-approved chemotherapeutics for the treatment of myelodysplastic syndromes and myeloid leukemia, while other DNMTis, including guadecitabine, hydralazine, and the green tea extract epigallocatechin gallate, are in clinical trials for various cancer types.
RNA Methylation and Disease Progression
In addition to pathological changes in DNA methylation, the link between altered epitranscriptomics and disease progression has been gaining attention. N6-methyladenosine (m6A) is the most prevalent modification in eukaryotic RNA and, concomitantly, the most widely investigated. Tightly regulated by the precise interplay between methylase “writers”, demethylase “erasers”, and binding protein “readers”, m6A affects virtually every facet of RNA biology, including structure, splicing, localization, translation, stability, and turnover. Aberrant expression of methylating and demethylating enzymes impacting m6A deposition, as well as of the m6A readers modulating RNA fate, have been implicated in diverse cancer outcomes, thus presenting potential targets for further medicinal exploration.
Tools for Epigenetic Research
Drug discovery and development would undoubtedly benefit from simple, quick, and reliable approaches to accurately measure DNA and RNA methylation in a high-throughput and cost-effective manner. Researchers have an assortment of techniques at their disposal to examine these epigenetic modifications at the genome-wide, gene-specific, and single-base resolution levels. Such methods generally rely on the use of antibodies that capture modifications of interest. Enzyme-linked immunosorbent assays (ELISAs) are a convenient way to rapidly assess DNA and RNA methylation globally before pursuing more in-depth and costly applications like qPCR or NGS.
As pioneers in the R&D of epigenetics-based research products, EpigenTek has leveraged its proprietary MethylFlash™ and EpiQuik™ technologies to develop robust and highly cited immunoassays for global DNA and RNA methylation analysis. The MethylFlash™ Global DNA Methylation (5-mC) ELISA Easy Kit and the EpiQuik™ m6A RNA Methylation Quantification Kit are complete sets of optimized buffers and reagents for the colorimetric quantification of 5mC DNA and m6A RNA. By combining the convenience and speed of ELISA with high sensitivity and specificity, these kits are valuable tools for any researcher studying DNA and RNA methylation in both physiological and disease contexts.




 Cart (0)
Cart (0)