Epigenetics is the study of heritable changes in gene function that do not involve alterations to the DNA sequence itself. These changes can be influenced by diverse mechanisms, including DNA methylation, RNA methylation, and histone modification. One of the key players in this field is DNA methylation, a chemical modification of DNA that plays a pivotal role in regulating gene expression, cellular differentiation, development, and disease. DNA methylation involves the addition of a methyl group to the cytosine (C) base of DNA, typically occurring at cytosine-guanine (CpG) dinucleotide sites. Methylation can silence gene expression by preventing the binding of transcription factors and other regulatory proteins to the DNA, effectively turning genes off.

How Bisulfite Sequencing Works
Among the assortment of methods employed by epigenetic researchers, bisulfite sequencing stands out as a powerful and widely-used technique for examining DNA methylation. Bisulfite sequencing enables the assessment of DNA methylation patterns with high precision at the single CpG site level. The technique typically involves the following steps:
- DNA Denaturation: The DNA sample is heated, causing it to separate into single strands, making it accessible for further treatment.
- Sodium Bisulfite Treatment: The denatured DNA is exposed to sodium bisulfite, which chemically converts unmethylated C’s to uracils (U’s), while leaving methylated cytosines (5-mC’s) unaltered.
- PCR Amplification: Using polymerase chain reaction (PCR), the treated DNA is amplified to create a pool of DNA fragments, some of which will contain C’s and others containing U’s.
- Sequencing: The amplified DNA is subjected to next-generation sequencing, which allows the identification of CpG sites that were originally methylated (5-mC) and those that were unmethylated (C). Advanced bioinformatics tools are then used to analyze the sequencing data and determine the methylation status of specific CpG sites.
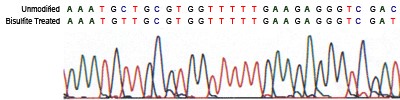
What to Watch Out For
To effectively and efficiently prepare bisulfite-converted DNA for use in various downstream analyses, an ideal DNA bisulfite modification method should be: 1) Highly accurate to allow for the complete conversion of unmethylated C to U (correct conversion) without deamination of 5-mC to thymine (inappropriate conversion); and 2) Fast enough to enable the bisulfite process to be as short as possible, since rapid DNA methylation analysis is highly demanded for basic research and, in particular, for clinical applications.
While bisulfite sequencing is a valuable tool, it does have its limitations. The traditional bisulfite conversion method needs 12-16 hours for bisulfite treatment, resulting in heavy DNA degradation (>80%), which can limit the amount and quality of DNA available for sequencing, and high inappropriate 5-mC deamination (>3.5%) and low cytosine conversion rate (<95%), which can lead to potential misinterpretation of methylation status.
Refining the Process
To overcome these limitations and enhance the effectiveness of bisulfite sequencing, EpigenTek developed the BisulFlash™ DNA Modification Kit. With its novel and optimized bisulfite composition, this kit shortens the entire bisulfite process from 16 hours to just 20 minutes, significantly improving cytosine conversion efficiency and effectively preventing converted DNA degradation. The advantages of this kit include:
- Speed: The entire procedure is reduced to as little as 30 minutes, without any reagent setup time.
- Efficiency: Unmethylated C is completely converted into U (modified DNA >99.9%).
- DNA Protection: More than 90% of DNA loss can be prevented, allowing for greater recovery.
- Sensitivity: Extremely low amount of input DNA is required for modification (only 0.2 ng or just 50 cells), making it ideal for precious or low abundance samples.
Converted DNA prepared with the BisulFlash™ DNA Modification Kit is suitable for a variety of post-bisulfite-based applications, such as methylation-specific PCR (MS-PCR), real-time MS-PCR, methylation microarray, and bisulfite-sequencing, including pyrosequencing and deep-sequencing.



 Cart (0)
Cart (0)