Chromatin immunoprecipitation (ChIP) offers an advantageous tool for studying protein-DNA interactions and can be used to determine whether a specific protein binds to a particular sequence of a gene, such as the target sequence of a transcription factor or to compare the levels of histone methylation associated with a specific gene promoter region between normal and diseased tissues. Identifying the genetic targets of DNA binding proteins and revealing the mechanism of protein-DNA interaction is crucial for understanding cellular processes.
Map genome-wide histone modifications with EpiGentek’s comprehensive chromatin immunoprecipitation sequencing (ChIP-Seq) platform. Optimized buffers and protocol allow minimal ChIP background and increased sensitivity and specificity of the ChIP reaction.
- All kits used in our ChIP-Seq services are commercially available with our ChIP-Seq Workflow.
- Highly efficient enrichment of targeted DNA. Enrichment ratio of positive to negative control > 500.
- High sensitivity and flexibility allows both non-barcoded (singleplexed) and barcoded (multiplexed) DNA library preparation.
- Delivery of high-quality publishable data.
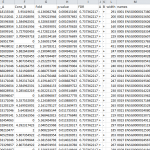
- Receive full deliverable package including Illumina FASTQ raw sequencing files, FastQC quality control insight, Bismark read mapping, Peak calling, peak annotation and differential binding analysis.
Example Data
*These are examples of data that may be provided. Please consult your service representative regarding the exact data you will receive.
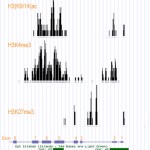
- A ChIP-sequencing library was prepared using the EpiNext ChIP-Seq High Sensitivity Kit (Cat. #P-2030) from rat heart chromatin and polyclonal antibodies against H3K4me3 (Cat. #A-4033), H3(K9/14)ac (Cat. #A-4021) and H3K27me3 (Cat #A-4039). Sequencing was carried out on an Illumina HiSeq2500. Bioinformatics analysis of ChIP-seq is performed utilizing Bowtie and MACS. ChIP-seq reveals that the peaks of H3K4me3 and H3(K9/14)ac, which are indicators of gene activation presence in the exon CpG islands (CGI) of transcription factor gene Nr1h2 (nuclear orphan receptor member), while H3K27me3, which causes gene silencing, is not enriched in the same gene. It is reported that H3K4me3 is often enriched in CGI of orphan genes.
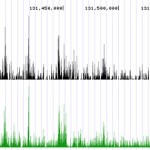
- ENCODE Quality ChIP-Seq Services. Histone H3K18ac ChIP-Seq reads provided by EpiGentek’s ChIP-Seq service align with the same peak sites as ENCODE data.
- Differential Binding Analysis includes peak annotation data and identifies where specific protein-DNA and transcription factor binding occurs.
Sample Requirements

We currently accept chromatin or ChIPed DNA for our ChIP Sequencing (ChIP-Seq) service. Proper sample preparation along with the appropriate quality control methods on your end will allow the greatest chance for assay success. Please follow all guidelines for sample requirements, packaging and labeling, and shipping which are outlined on the Submission Instructions page.
Talk to a Scientist

Want more information on our ChIP Sequencing (ChIP-Seq) service or interested in customizing your research project? We’re here to help! Get in touch with one of our highly knowledgeable epigenetic services experts by calling toll-free at 1-877-374-4368 or emailing services @ epigentek.com.